FISHBOOST January Newsletter

This newsletter updates you on recent developments in aquaculture breeding and the FISHBOOST project.
This is FISHBOOST:
FISHBOOST is an FP7 EU resesarch project that works on improving the main components of breeding programmes for the main six fish species produced in Europe:
Atlantic salmon, common carp, European seabass, gilthead seabream, rainbow trout and turbot.
In this edition:
- FISHBOOST is finalizing!
- FISHBOOST announcements
- The role of FISHBOOST in research-oriented European education process
- Breeding programmes prove their worth
- The importance of ensuring genetic variability when establishing aquaculture breeding programmes
- Use of DNA pooling of the training set to reduce genotyping costs for genomic selection
- Two well-visited FISHBOOST sessions at European Aquaculture Society meeting
- In the spotlight: Andrea Doeschl-Wilson and Santiago Cabaleiro Martinez
______
FISHBOOST is finalizing!

FISHBOOST had its final meeting in Brussels with an open session to policymakers and other stakeholders on January 22-23, 2019.
While the first day focused on the progress since last Annual Meeting, second day focused on the impact FISHBOOST created in the European aquaculture breeding and production sector. Following the introduction of the main activities and research in FISHBOOST, the industry stakeholders in FISHBOOSTSYSAAF, Salmobreed and Klatryb were given the floor to explain their role in FISHBOOST and FISHBOOST impact in their companies.
The audience also had the chance to view the FISHBOOST Impact movie produced by NOFIMA during the meeting. Results of the survey and round table discussions during Farmer’s Day event and Advanced Breeding Training were presented by Anna Sonesson, the FISHBOOST Coordinator. The meeting was closed with the Horizon 2020 presentation of Marta Iglesias from the European Commission and the demonstration of the “Aquaculture App: by Francois Allal from Ifremer.
FISHBOOST announcements
Carp : 219 SNPs
Rainbow Trout : 1048 SNPsThis chip will be used for the Fishboost project purposes, and Labogena will be glad to discuss with you if you have any interest for any other use, e.g. add Sea-Bass SNPs or other species if there is any interest. For further discussion, please contact Julien Pradelles.
The role of FISHBOOST in research-oriented European education process
Martin Prchal (USB)
FISHBOOST is a European research project that benefits from collaborations between top research European institutions focused on animal breeding. Within the various research groups, PhD students work on their dissertation topics. Kasper Janssen, PhD candidate at Wageningen University, department of Animal Breeding and Genetics, works on the economic evaluation of breeding programs, and Clémence Fraslin, PhD candidate at INRA (Jouy-en-Josas, France) in collaboration with the SYSAAF (French Poultry and Aquaculture Breeders Association), aims to investigate the genetic architecture of rainbow trout response to Flavobacterium psychrophilum, the causative agent of Bacterial Cold Water Disease. Clémence will defend her work on December 20th in Paris. Recently, FISHBOOST’s first PhD candidate, Martin Prchal, defended his thesis.
Martin was a Ph.D. student at the University of South Bohemia in České Budějovice (USB), Czech Republic. His work focussed on common carp breeding. Martin contributed to four research papers within the FISHBOOST project, either as first author or co-author. Martin is the first PhD student from FISHBOOST that defended his PhD thesis and is entitled “Estimation of genetic variation of performance traits in common carp to predict potential of selective breeding under pond management conditions”. The defence was on 20th September 2018 and can be viewed online:http://alpha.frov.jcu.cz/eduart/frov/2018/phd_def/03_prchal/.
During the study, Martin had the opportunity to collaborate with FISHBOOST partners to mutually share their knowledge, skills and infrastructure. Martin also trained his presentation skills when informing and discussing about plans and results on the annual general assemblies of the FISHBOOST project. Moreover, he stayed one month abroad, for collaboration with the Natural Resources Institute (LUKE), Jokioinen in Finland, one of the FISHBOOST partners. Under supervision of Dr. Antti Kause, he learnt more about the quantitative genetics of fish, and especially about estimation of genetic parameters and possibilities to set up the selective breeding program in common carp. Due to this close collaboration with experts from various fields, solid results could be published in highly valued journals.
Common carp is a highly important fish species in European aquaculture. Yet, genetic improvement via selective breeding has not been developed. The thesis of Martin investigated the use of modern selection methods in genetic improvement of e.g. growth or slaughter yields, and showed there is great potential. Base populations of common carp have recently been established in cooperation with the Klatovy fish farm, another FISHBOOST partner. Moreover, the first selection challenge for faster growth was applied. Hence, FISHBOOST has significantly contributed to knowledge that supports the importance of selective breeding in common carp, and also fulfilled the main aim related to the shifting of selective breeding in this species to the first level.
Breeding programmes prove their worth
Rob Fletcher (orginally published at The Fish Site)
The benefits that salmon farmers obtain by sourcing their stocks from cutting-edge breeding programmes are chronically undervalued, according to a new study. The study, ‘Cost-benefit analysis of a breeding program for Atlantic salmon’, estimated the economic impacts of genetic improvement, focusing on traits such as improved growth rates, lower FCRs, better fillet yields and improved resistance to sea lice. Results were obtained by simulation of a breeding programme that was based on the breeding programme used by SalmoBreed prior to 2016.
The results of the project were presented to delegates at Aqua2018 in Montpellier by Kasper Janssen. “The aim is to quantify the benefits that farmers gain from genetic improvement every generation – both by traditional family breeding programmes and by the use of genomic selection – as the economic benefits that producers get from the breeding companies hasn’t been effectively calculated before,” explained Kasper Janssen, from Wageningen University, who presented the research at Aqua2018 in Montpellier last week.
The first part of the study, which calculated the economic impacts of a generation produced by family selection where the emphasis on breeding goal traits is at its economic optimum, found that the combined effect of improving growth and FCR would increase profit by €245 per tonne of salmon produced per generation, and a saving in terms of lice treatments of €8 per tonne of salmon produced, by reducing the average treatments per cycle from 3.7 to 3.6. Overall, according to the study, using a strain produced by family selection led to a small increase in production volumes as well as cost savings, resulting in a profit increase of €275/tonne per generation.
The study forecasts that the annual genetic gain should equate to €50-80 per tonne of salmon produced per year. The second part of the study looked at the economic impact of a generation produced by genomic selection and the results suggest an even higher increase in profit for the producers, with growth rate up an additional 4 percent, FCR down 8 percent more, sea lice resistance up an extra 9 percent and fillet yield up 4 percent – leading to a €291/tonne benefit for the farmer per generation (every 3.7 years).
Moreover, the study forecasts that the annual genetic gain should equate to €50-80 per tonne per year. And, given that each egg leads to an average yield of 3.8 kg, the genetic potential of an egg would thereby increase by €0.20-0.30 per year, despite the sales price of each egg being less than €0.20 and typically increasing by only ~€0.01 per year Whether the rate of economic improvement per generation will be linear remains to be seen.
As Janssen reflects: “I expect that such results can be sustained over generations, although they become more uncertain towards the future. As long as genetic diversity is maintained by controlling the rate of inbreeding (which all breeding companies should), trait levels will be improved by selection. Their effect on farm profit may however be non-linear, such that future economic gains may change. On the other hand, gains may even increase with improvements in genomic techniques.”
He also adds the caveat that his study was not able to take all variables into account. Despite this, however, he feels that the underlying conclusions of the research are both striking and sound.
The importance of ensuring genetic variability when establishing aquaculture breeding programmes
María Saura (INIA, Spain)
Designing base populations with appropriate levels of genetic variability is fundamental to ensure the viability of aquaculture breeding programmes. Using genomic data obtained within FISHBOOST, INIA has determined the genetic status of commercial populations of turbot, gilthead seabream, European seabass and common carp, and shown the need of broadening the genetic composition of base populations from which selection programmes start and the application of strategies for maintaining genetic diversity once the programmes are in operation.
Importance of optimising the creation of base populations
The success of any breeding programme critically depends on the way in which the base population of breeders is built, as the genetic variability initially available will determine the genetic progress achieved. This is particularly important in aquaculture since given the high fecundity of fish, base populations can be created from very few individuals, leading to large losses in genetic variability and increased risk of inbreeding depression (i.e. reduction of performance due to inbreeding) that can compromise the viability of the programmes.
Genetic variability and effective population size
The rate of loss of genetic variability is directly related to the rate at which inbreeding increases and inversely related with the effective population size, a population parameter that can be estimated from genomic information. Using data obtained within FISHBOOST, INIA has estimated current and ancestral effective population size in populations of turbot, gilthead seabream, European seabass and common carp. Data used came from different challenge tests carried out in the project and corresponded to the broodstock used for creating the families challenged. These broodstock were sampled from different commercial European breeding programmes. Turbot data were obtained from the broodstock maintained at CETGA (Aquaculture Cluster of Galicia, Spain), a population that is representative of the main European breeding programmes that have the same Atlantic origin. Seabream data came from two different breeding programmes carried out at Ferme Marine du Douhet (FMD, France) and the Andromeda Group (Greece). Carp data came from the broodstock of the Amur Mirror Carp strain, recently created at the University of South Bohemia (Czech Republic) from crosses between two different strains. Fish were genotyped for thousands of Single Nucleotide Polymorphisms (SNPs) obtained using genotyping-by-sequencing (RAD-sequencing), and estimates of current and ancestral effective population size from linkage disequilibrium measures were obtained.
Genetic status of turbot, seabream, seabass and carp commercial populations
Results obtained from the analysis of genomic information revealed that current effective population size for all populations analysed was lower than the critical value of 50 individuals that is recommended to avoid inbreeding depression and retain fitness in the short-term. Specifically, estimates for turbot, seabream Andromeda, seabream FMD, seabass and carp were 28, 40, 41, 38 and 22 fish, respectively. These low estimates could be due to the way in which base populations were established (low number of breeding individuals and/or strong relationships between them) or/and to a suboptimal inbreeding control. Estimates of effective population size across time evidenced bottlenecks 4 – 11 generations ago. This point in time coincides with the number of generations for which turbot, seabream and seabass have been domesticated or for which selection programmes have been practised. The recent creation of the carp strain analysed also coincides with the observed bottleneck.
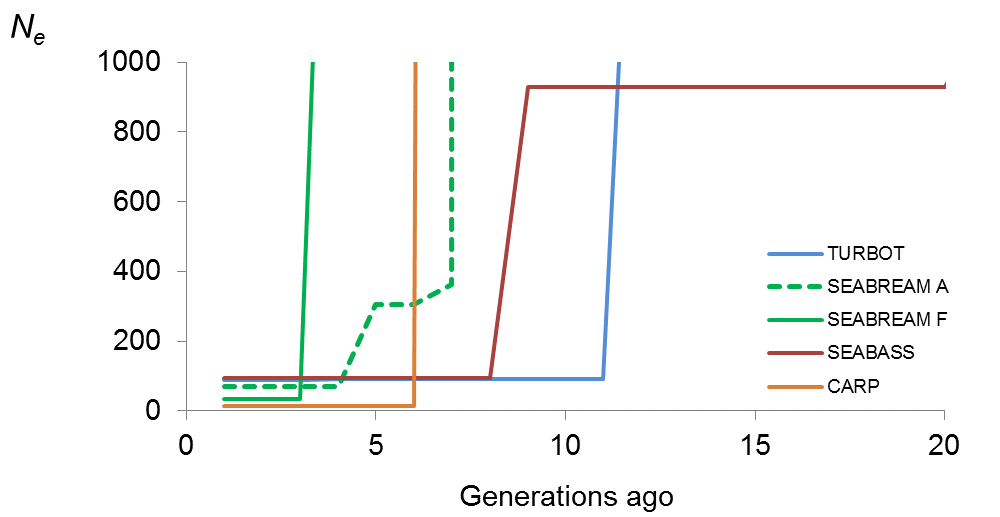
Estimates of effective population size (Ne) across the last 20 generations for the different populations analysed
Implications
Our results highlight the need of broadening the genetic composition of base populations from which selection programmes start. These results also suggest that methods designed to increase the effective population size within all farm populations analysed, need to be implemented in order to ensure the sustainability of the breeding programmes. Performing artificial fertilization and single-pair matings as well as implementing Optimal Contribution Selection and factorial mating designs would be recommendable practices
This study was performed by M. Saura, A. Fernández, J. Fernández and B. Villanueva (INIA). Software for estimating effective population size was facilitated by A. Caballero (Universidad de Vigo, Spain) and E. Santiago (Universidad de Oviedo, Spain). Data analysed was provided by CETGA (S. Cabaleiro), GENEAQUA (P. Martínez, A. Millán), USB (M. Kocour, M. Prchal), UEDIN (C. Palaiokostas, Ross Houston), ANDROMEDA (T. Kostas), NOFIMA (M.L. Aslam), UNIPD (L. Bargelloni), SYSAAF (P. Haffray) and FMD (J.S. Bruant).
Use of DNA pooling of the training set to reduce genotyping costs for genomic selection
Anna Kristina Sonesson (NOFIMA)
 selection is mainly used for traits that are not measured on the selection candidates, because they are invasive, but instead they are measured on sibs of the selection candidates. Examples of these are disease resistance and fillet quality, and they often constitute a large group of the traits in the breeding goals of well-developed breeding programs. Phenotypic and genomic information must be obtained from the training set of individuals, i.e. the sibs of the candidates. The effects of the SNPs estimated in this dataset are thereafter used to predict genomic breeding values of the selection candidates. With large numbers of traits and individuals per trait (15-50 per family), the total number of individuals to genotype becomes large. Methods that reduce genotyping costs for genomic selection are therefore important for genomic selection to be widely implemented in aquaculture breeding schemes.
selection is mainly used for traits that are not measured on the selection candidates, because they are invasive, but instead they are measured on sibs of the selection candidates. Examples of these are disease resistance and fillet quality, and they often constitute a large group of the traits in the breeding goals of well-developed breeding programs. Phenotypic and genomic information must be obtained from the training set of individuals, i.e. the sibs of the candidates. The effects of the SNPs estimated in this dataset are thereafter used to predict genomic breeding values of the selection candidates. With large numbers of traits and individuals per trait (15-50 per family), the total number of individuals to genotype becomes large. Methods that reduce genotyping costs for genomic selection are therefore important for genomic selection to be widely implemented in aquaculture breeding schemes.
In a smaller dataset of Atlantic salmon, also tested for PD, and individually genotyped with a SNP chip, the selection accuracy was 0.737. DNA from these same individuals were in vivo pooled into two replicates of SURVIVORS and MORTALITIES pools, each with ~200 individuals from ~30 families. When these pools were sequenced 40x, selection accuracy was 0.716. When these pools were genotyped with a SNP chip and relative light intensities were used to estimate SNP alleles frequencies in the pools, the accuracy of selection was 0.690.
In conclusion, genotyping costs of genomic selection can be dramatically reduced when performing DNA pooling of extreme phenotypes of the training population. Selection accuracies were slightly reduced compared to individual genotyping, but this reduction became smaller when the number of pools increased. The methodology worked best in large populations.
This work was done in collaboration between SalmoBreed AS, Marine Harvest, Nofima, Norwegian University of Life Sciences. Please, contactanna.sonesson@nofima.no if you have questions.
Photo credits to Frank Gregersen and Nofima.
Two well-visited FISHBOOST sessions at European Aquaculture Society meeting
This years’ European Aquaculture Society meeting was co-organised with the World Aquaculture Society and was held in Montpellier, France, in the end of August. FISHBOOST had two very well-visited sessions at this conference.
One session was part of the Industry Forum, where industry relevance is in focus. Overviews of the FISHBOOST results so far were presented. The covered topics were improvement of disease and production efficiency traits, genetic gains and profitability of breeding programs, management of inbreeding and genomic selection. At the end of that session, Dr. Nikos Zampoukas from the European Commission was invited to present the new Framework Program, Horizon Europe.
![]() This was followed up by a round table discussion on the future research needs within the aquaculture breeding field. The results from that discussion will be summarized and sent as input for future calls to the European Commission via the Farm Animal Breeding and Reproduction Technology Platform (FABRE TP). You can read more about the FABRE TP here: http://www.fabretp.eu/
This was followed up by a round table discussion on the future research needs within the aquaculture breeding field. The results from that discussion will be summarized and sent as input for future calls to the European Commission via the Farm Animal Breeding and Reproduction Technology Platform (FABRE TP). You can read more about the FABRE TP here: http://www.fabretp.eu/
The second session was a technical session with eight presentations on the latest results from the project.
- The importance of ensuring genetic variability when establishing selection programmes in aquaculture. B. Villanueva, M. Saura, A. Caballero, E. Santiago, E. Morales, A. Fernández, J. Fernández, S. Cabaleiro, P. Martínez, A. Millán, C. Palaiokostas, M. Kocour, R. Houston, M. Prchal, L. Bargelloni, K.Tzokas.
- Genomic selection analyses results on common carp, European seabass, gilthead beabream and turbot. A.K. Sonesson, C. Palaiokostas, R.D. Houston, B. Dagnachew, M.L. Aslam, T.H.E. Meuwissen.
- Mapping, sequencing and functional annotation of a QTL region affecting resistance to koi herpesvirus in common carp. R.D. Houston, C. Palaiokostas, D. Robledo, T. Vesely, M. Prchal, D. Pokorova, V. Piackova, L. Pojezdal, M. Kocour
- A closer look at turbot genome reveals a genetic component of parasite resistance: new tools for selection. P. Martínez, F. Maroso, M. Saura, A. Fernández, A. Blanco, M. Hermida, S. Cabaleiro, A. Doeschl-Wilson, O. Anacleto, R. Houston, A. Millán, J. Fernández, L. Bargelloni, G. dalla Rovere, M.A. Toro, M.J. Carabaño, C. Bouza, B. Villanueva.
- Use of DNA pooling in genomic selection for a disease trait in Atlantic salmon. B. Dagnachew, A. K. Sonesson, T.H.E. Meuwissen.
- Successful realized selection response for fillet yield in rainbow trout (Oncorhynchus mykiss). M. Vandeputte, J. Bugeon, A. Bestin, A. Desgranges, S. Courant, J-M Allamellou, A.S. Tyran, F. Allal, M. Dupont-Nivet, P. Haffray.
- Potential for genetic improvement of the main slaughter yields in common carp with in vivo morphological predictors.M. Prchal, J. Bugeon, M. Vandeputte, A. Kause, A. Vergnet, J. Zhao, D. Gela, L. Genestout, A. Bestin, P. Haffray, M. Kocour.
- Optimal schemes for advancing selective breeding to the next level for the main species of European aquaculture. B. Villanueva, S. García-Ballesteros, J. Fernández.
- Cost-benefit analysis of a breeding program for Atlantic salmon. K. Janssen, H. Saatkamp, H. Komen.
A summary of this session can be found at of www.aquaeas.eu.
Thanks to everybody that has visited the FISHBOOST sessions at this and previous EAS meetings for stimulating discussions!
In the spotlight:
Andrea Doeschl-Wilson (The Roslin Institute)
I’m contributing to the disease resistance workpackage (WP1). Specifically, we are looking at how the genetics of individuals affects disease spread and survival in fish populations. In addition to the conventional trait ‘disease resistance’, we consider new traits (disease phenotypes), such as tolerance and infectivity. We know from theory that all of these traits also have a strong influence on disease spread and resulting mortality rates.
However, at present, very little is known about the genetic regulation of these traits, and whether it is possible to breed fish that are not only genetically less prone to becoming infected, but also less likely to transmit infection once infected, and possibly also more likely to survive or recover from infections. If this was possible, genetic selection would be a powerful tool to prevent disease outbreaks – potentially much more powerful than selection on survival, which is the current practice. In FISHBOOST we develop the methods and know-how needed to estimate genetic effects for the different disease traits.
The Roslin Institute is one of the world leading research institute in livestock genomics and breeding, including aquaculture. Together with CETGA and INIA, we have designed a large scale disease transmission experiment for an important disease in Turbot (Scuticociliatosis) to generate data that allow us to simultaneously estimate genetic effects for resistance, tolerance in infectivity. This is the first experiment in the world that dissects the black box ‘mortality’ as indicator for disease resistance into its underlying components. If you are selecting fish that are genetically more prone to survive infection without knowing what underlies survival, you could accidentally select fish that are more tolerant to infection and act as source of infection to others. In FISHBOOST we are developing the experiment design and also the mathematical models and computational algorithms needed to estimate genetic effects for the new traits infectivity and tolerance, in addition to susceptibility. We apply them to the experimental data generated within FISHBOOST. We hope to apply these to other diseases in aquaculture species in the future.
I’m very impressed about the scope of this project and the immense dedication of the project leaders and partners to collaboratively carry out relevant research that will have important practical implications to the aquaculture sector. This project generates a number of important technological and statistical tools and know-how to move aquaculture production in Europe a significant step forward. I’m very grateful to be part of it.
 We could show that it is possible to conduct single transmission experiments that allows detection of genetic variation not only in mortality following a disease challenge, but also in all three important underlying host traits resistance, infectivity and tolerance. The data from the Turbot transmission experiments revealed that there is significant genetic variation in all of these traits, meaning that it would be possible in principle to include all three disease traits into a breeding programme. For Scuticocciliatosis in Turbot, resistance and tolerance appear to be genetically different traits (i.e. low genetic correlations), but resistance seems to be highly genetically correlated with mortality. Genome-wide association analyses carried out by our INIA collaborators also detected QTLs controlling tolerance and mortality. Although no significant associations were found for resistance, the pattern of association was the same as for mortality. The transmission experiment also provided the first empirical evidence that individuals not only differ genetically in their risk of becoming infected and how they cope with infection, but also in their ability to transmit infection (i.e. their infectivity). So far we could demonstrate that the genetic differences in infectivity not only affect disease spread but also survival rates. Analyses to estimate the relationship between infectivity and the other disease traits are currently under way.
We could show that it is possible to conduct single transmission experiments that allows detection of genetic variation not only in mortality following a disease challenge, but also in all three important underlying host traits resistance, infectivity and tolerance. The data from the Turbot transmission experiments revealed that there is significant genetic variation in all of these traits, meaning that it would be possible in principle to include all three disease traits into a breeding programme. For Scuticocciliatosis in Turbot, resistance and tolerance appear to be genetically different traits (i.e. low genetic correlations), but resistance seems to be highly genetically correlated with mortality. Genome-wide association analyses carried out by our INIA collaborators also detected QTLs controlling tolerance and mortality. Although no significant associations were found for resistance, the pattern of association was the same as for mortality. The transmission experiment also provided the first empirical evidence that individuals not only differ genetically in their risk of becoming infected and how they cope with infection, but also in their ability to transmit infection (i.e. their infectivity). So far we could demonstrate that the genetic differences in infectivity not only affect disease spread but also survival rates. Analyses to estimate the relationship between infectivity and the other disease traits are currently under way.
The results and tools generated in FISHBOOST could help breeders to develop more effective breeding programmes that can exploit genetic variation in several key host traits underlying disease spread and survival.
Breeding for disease resistance is currently still in it’s infancy. I believe that there are far more opportunities for genetic disease control, in particular in aquaculture, by optimising experimental and field study designs, by making better use of genomic technologies and developing better statistical models that combine genetic and epidemiological concepts. I hope that eventually these will be taken up by industry.
Santiago Cabaleiro Martinez (CETGA)
CETGA is involved in FISHBOOST concerning culturing all the families used for turbot challenges in all the Work Packages. 
As Research Centre, we developed most of the trials regarding turbot: detecting genetic parameters to improve feed efficiency, performing different disease challenges and recording mortalities, time-to-death and growth. The cooperation among all the participants allow us to obtain accurate data about the genetic breeding selection, establishing for the first time a correlation between resilience, tolerance and infectivity that will allow to improve the breeding selection process.
All the results obtained from this project are relevant to the stakeholders and this is what makes a project profitable. So, Aquaculture industry can improve growth rates, minimize mortality and enhance filet% taking into account the results of the project, These results establishes a closer  relationship with the stakeholders, we can improve their breeding selection thanks to new technologies and innovative procedures.
relationship with the stakeholders, we can improve their breeding selection thanks to new technologies and innovative procedures.
Moreover, our cluster includes the largest aquaculture producers in Spain and South Europe, which allows us to be directly in contact with the industry, what let us know about their concerns, difficulties and necessities. All the data obtained in research projects are transferred to the European industry.






